Difference between revisions of "Real-Time ECG Contractions Classification"
From iis-projects
| (28 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | [[File: | + | [[Category:Biomedical System on Chips]] |
| + | [[File:ecg_task.png|thumb|ECG Task]] | ||
==Introduction== | ==Introduction== | ||
| + | ECG (electrocardiography) is the process of recording the heart-generated electrical activity over a period of time using skin-placed electrodes. | ||
| + | A well defined sequence of atrial and ventricles contractions allow the blood to be pumped in the circulatory systems, essential to enable many physiological functions as well as life. Such recorded electrical activity reflects the underlying contractions of the heart and have well defined taxonomy (according to shapes, peaks, timing, etc), allowing to detect and label heart diseases. | ||
| − | + | Nowadays, ECG interpretation relies on eye inspection by trained medical doctors, with expensive and time-consuming manual analysis. Patient-to-patient variability, different recording conditions and settings and patient clinical conditions makes the automation of this task very challenging. | |
| + | For this reason the ECG monitoring is currently limited to short-time scales (minutes to hours) in well controlled environments and setups (clinics, expensive devices). | ||
| − | + | Even though current ECG exams works well if there is already the suspect of a heart disease going on, this prevents to detect outliers in the ECG, i.e., abnormal contractions that rarely happens and the chances to observe them under clinical ECG are very low. | |
| − | + | Similarly, drifting over time of ECG parameters may not be noticed with normal ECG acquisitions, as the time scale is very short. | |
| + | As a result, many potential diseases can't be early-detected due to these limitations. Technology advancement in hardware design and in data-driven algorithms give a hope providing tools to change ECG detection, from expensive non-routine exams, to cheap daily monitoring. | ||
==Short Description== | ==Short Description== | ||
| − | Your mission, should you choose to accept it, is to join our research into | + | Your mission, should you choose to accept it, is to join our active research into biomedical system design. The approach is to used state-of-the-art machine learning algorithms to classify different ECG contractions. Particular emphasis will be devoted to detect outliers in the ECG, which usually are not even noticed by the patient, but by which are known to anticipated important disease if not timely cured [1]. Given the constrained conditions under which we operate, i.e, wearable devices, energy-efficiency is of paramount importance. |
| − | ===Status: | + | The starting point is the VivoSoC platform, a system on chip capable to acquire and process ECG signals. An open-source labelled ECG dataset is available online ready to be used [2][3]. |
| − | : | + | |
| − | : | + | The task includes the following main sub-points: |
| + | <ul><li> Understand the ECG basics and interpret the dataset.</li> | ||
| + | <li> Develop (high-level Phython or Matlab) a supervised-learning classification algorithm to classify the ECG contractions. </li> | ||
| + | <li> Map the algorithm in the VivoSoC hardware (C-programming PULP).</li> | ||
| + | <li> Conduct in-vivo experiments to validate the method with a realistic setting.</li></ul> | ||
| + | |||
| + | The task is anyway flexible and it will be adapted the student skills and will. | ||
| + | |||
| + | |||
| + | ===Status: Completed === | ||
| + | : Semester Project in Fall 2018 (Sumu Zhao, sem18h16) | ||
| + | : Supervised by: [[:User:Glaserf | Florian Glaser]], [[:User:xiaywang|Xiaying Wang]] | ||
<!-- | <!-- | ||
===Status: Completed === | ===Status: Completed === | ||
| Line 27: | Line 43: | ||
===Character=== | ===Character=== | ||
| − | : | + | : 40% Theory and Algorithms |
| − | : | + | : 40% Implementation (C coding) |
| − | : | + | : 20% Verification and Testing |
===Prerequisites=== | ===Prerequisites=== | ||
| − | + | : Knowledge in Machine Learning, from preprocessing, feature extraction, classifier, supervised-learning | |
| − | + | : Embedded system programming | |
| − | + | : Basic analog / ADC / sampling theory hands-on knowledge. | |
| − | |||
| − | |||
| − | |||
| − | : Knowledge in Machine Learning, | ||
: C/C++ | : C/C++ | ||
===Professor=== | ===Professor=== | ||
: [http://www.iis.ee.ethz.ch/people/person-detail.html?persid=194234 Luca Benini] | : [http://www.iis.ee.ethz.ch/people/person-detail.html?persid=194234 Luca Benini] | ||
| − | |||
===References=== | ===References=== | ||
| − | : [1] | + | : [1] B.A.Koplanand et al, “Ventricular Tachycardia and Sudden Cardiac Death,” Elsevier Mayo Clinic Proceedings, vol. 84, no. 3, pp. 289–297, 2009. |
| − | + | : [2] https://www.physionet.org/physiobank/database/mitdb/ | |
| − | + | : [3] https://physionet.org/lightwave/ | |
| − | : [ | ||
| − | : [ | ||
| − | |||
| − | |||
[[#top|↑ top]] | [[#top|↑ top]] | ||
| − | + | ||
[[Category:Digital]] | [[Category:Digital]] | ||
| − | [[Category: | + | [[Category:Analog]] |
| − | |||
[[Category:Semester Thesis]] | [[Category:Semester Thesis]] | ||
[[Category:Master Thesis]] | [[Category:Master Thesis]] | ||
| + | [[Category:Completed]] | ||
| + | [[Category:Glaserf]] | ||
| + | [[Category:Xiaywang]] | ||
| + | [[Category:Rovereg]] | ||
[[Category:PULP]] | [[Category:PULP]] | ||
| − | |||
| − | |||
| − | |||
[[Category:Software]] | [[Category:Software]] | ||
| + | [[Category:Processor]] | ||
| + | [[Category:Biomedical System on Chips]] | ||
Latest revision as of 19:15, 9 March 2020
Contents
Introduction
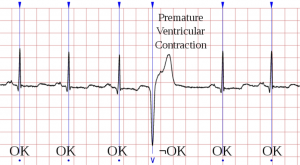
ECG (electrocardiography) is the process of recording the heart-generated electrical activity over a period of time using skin-placed electrodes. A well defined sequence of atrial and ventricles contractions allow the blood to be pumped in the circulatory systems, essential to enable many physiological functions as well as life. Such recorded electrical activity reflects the underlying contractions of the heart and have well defined taxonomy (according to shapes, peaks, timing, etc), allowing to detect and label heart diseases.
Nowadays, ECG interpretation relies on eye inspection by trained medical doctors, with expensive and time-consuming manual analysis. Patient-to-patient variability, different recording conditions and settings and patient clinical conditions makes the automation of this task very challenging. For this reason the ECG monitoring is currently limited to short-time scales (minutes to hours) in well controlled environments and setups (clinics, expensive devices).
Even though current ECG exams works well if there is already the suspect of a heart disease going on, this prevents to detect outliers in the ECG, i.e., abnormal contractions that rarely happens and the chances to observe them under clinical ECG are very low. Similarly, drifting over time of ECG parameters may not be noticed with normal ECG acquisitions, as the time scale is very short. As a result, many potential diseases can't be early-detected due to these limitations. Technology advancement in hardware design and in data-driven algorithms give a hope providing tools to change ECG detection, from expensive non-routine exams, to cheap daily monitoring.
Short Description
Your mission, should you choose to accept it, is to join our active research into biomedical system design. The approach is to used state-of-the-art machine learning algorithms to classify different ECG contractions. Particular emphasis will be devoted to detect outliers in the ECG, which usually are not even noticed by the patient, but by which are known to anticipated important disease if not timely cured [1]. Given the constrained conditions under which we operate, i.e, wearable devices, energy-efficiency is of paramount importance.
The starting point is the VivoSoC platform, a system on chip capable to acquire and process ECG signals. An open-source labelled ECG dataset is available online ready to be used [2][3].
The task includes the following main sub-points:
- Understand the ECG basics and interpret the dataset.
- Develop (high-level Phython or Matlab) a supervised-learning classification algorithm to classify the ECG contractions.
- Map the algorithm in the VivoSoC hardware (C-programming PULP).
- Conduct in-vivo experiments to validate the method with a realistic setting.
The task is anyway flexible and it will be adapted the student skills and will.
Status: Completed
- Semester Project in Fall 2018 (Sumu Zhao, sem18h16)
- Supervised by: Florian Glaser, Xiaying Wang
Character
- 40% Theory and Algorithms
- 40% Implementation (C coding)
- 20% Verification and Testing
Prerequisites
- Knowledge in Machine Learning, from preprocessing, feature extraction, classifier, supervised-learning
- Embedded system programming
- Basic analog / ADC / sampling theory hands-on knowledge.
- C/C++
Professor
References
- [1] B.A.Koplanand et al, “Ventricular Tachycardia and Sudden Cardiac Death,” Elsevier Mayo Clinic Proceedings, vol. 84, no. 3, pp. 289–297, 2009.
- [2] https://www.physionet.org/physiobank/database/mitdb/
- [3] https://physionet.org/lightwave/